Task creation instruction
I. General requirements and procedure
- The program must be an executable file (compiled),
- if using additional non-standard libraries, they must be compiled statically with the program,
- Input and output files must have symbolic names (e.g. in0, in1, in2,out0,out1, ....),
- A list of symbolic names must be provided when creating the task,
- The application should be compiled with glibc version 2.28,
- The program must have the filename "Gaia@home_application".
II. Scientific problem
Task:
- Do a cone search on the Pleiades and retrieve pmra and pmdec for all stars from Gaia archive:
SELECT pmra, pmdec FROM gaiadr3.gaia_source_lite WHERE 1 = CONTAINS(POINT(56.75, 24.12),CIRCLE(ra,dec,2.0)) AND ruwe <1.4 - Make a further downselection by requiring that (pmra-(20))^2 + (pmdec-(-45))^2 < 5^2
- Compute the average pmra and pmdec for this subset using numpy
- Compute the standard deviation for this subset w.r.t. the average using numpy
- Write these four numbers to output file.
Python code for one CPU program:
#PLEIADES - one CPU program
#Searching the Pleiades and computing the mean value and standard deviation of proper motion
import numpy as np
#launching asynchronous job using astroquery
job = Gaia.launch_job_async(query="select pmra,pmdec "
"from gaiadr3.gaia_source_lite "
"where 1 = contains(point(56.75, 34.12),circle(ra, dec, 2.0)) "
"and ruwe<1.4 ")
r = job.get_results()
#constants are defined in an input config file
with open("Symbolic_config") as f:
line = f.readlines()[0].split(' ')
pmra0=float(line[0])
pmdec0=float(line[1])
#filtering the output from Gaia archive for objects with (pmra-pmra0)**2+(pmdec-pmdec0**2) < 5**2
#and saving results to numpy arrays
pm_ra=np.array([])
pm_dec=np.array([])
for row in r:
if (row["pmra"]-pmra0)**2+(row["pmdec"]-pmdec0)**2 < 25.0:
pm_ra=np.append(pm_ra,row["pmra"])
pm_dec=np.append(pm_dec,row["pmdec"])
#computing mean values and standard deviation using numpy
mean_pmra=np.mean(pm_ra)
mean_pmdec=np.mean(pm_dec)
stdev_pmra=np.std(pm_ra)
stdev_pmdec=np.std(pm_dec)
#saving results to output file
with open("Symbolic_result",'a') as f:
f.write(f'{mean_pmra},{mean_pmdec}\n{stdev_pmra},{stdev_pmdec}\n')
Input file:
config.inp
20
-45
III. Paralleling the issue
We perform task parallelization at the level of the query to the Gaia archive.
In our example, a single database query
SELECT pmra, pmdec FROM gaiadr3.gaia_source_lite WHERE 1 = CONTAINS(POINT(56.75, 24.12),CIRCLE(ra,dec,2.0)) AND ruwe <1.4
is replaced by a sequence of queries, dividing the searched area into smaller fragments of a given size.
loop (RA_min<RA<RA_max with step RA_step)
loop(DEC_min<DEC<DEC_max with step DEC_step)
SELECT pmra, pmdec FROM gaiadr3.gaia_source_lite WHERE 1 = CONTAINS(POINT(ra,dec),BOX(RA,DEC,RA_step,DEC_step)) AND ruwe <1.4 Queries are executed by gaia@home service and written to the input file for each task individually.
In our example, the values will be as follows:
RA_min = 56.75 - 2 = 54.75 deg = 03:39:00
RA_max = 56.75 + 2 = 58.75 deg = 03:55:00
DEC_min= 24.12 - 2 = 22.12 deg = 22:07:12
DEC_max= 24.12 + 2 = 26.12 deg = 26:07:12
RA_step and DEC_step - size of each small area (e.q. 5 marcsec)
Assume that these files have the name gaia_data.inp
Our program prepared for parallel operation looks as follows:
#PLEIADES - parralel with phisical name of files
#Searching the Pleiades and computing the mean value and standard deviation of proper motion
import numpy as np
#loading gaia results (using numpy to read whole table)
#use defined symbolic name as file names when opening the files
r=np.loadtxt(fname="gaia_data.inp", skiprows=1)
#constants are defined in an input config file
with open("config.inp") as f:
line = f.readlines()[0].split(' ')
pmra0=float(line[0])
pmdec0=float(line[1])
#filtering the output from Gaia archive for objects with (pmra-pmra0)**2+(pmdec-pmdec0**2) < 5**2
#and saving results to numpy arrays
pm_ra=np.array([])
pm_dec=np.array([])
for row in r:
if (row[0]-pmra0)**2+(row[1]-pmdec0)**2 < 25.0:
pm_ra=np.append(pm_ra,row[0])
pm_dec=np.append(pm_dec,row[1])
#computing mean values and standard deviation using numpy
mean_pmra=np.mean(pm_ra)
mean_pmdec=np.mean(pm_dec)
stdev_pmra=np.std(pm_ra)
stdev_pmdec=np.std(pm_dec)
#saving results to output file
with open("result.dat",'a') as f:
f.write(f'{mean_pmra},{mean_pmdec}\n{stdev_pmra},{stdev_pmdec}\n')
The final step is to link the names of the input and output files to the symbolic names required by the BOINC system.
Symbolic names in BOINC can be arbitrary but must, at task runtime, be associated with passed input files.
Code: BOINC:
input.inp Symbolic_input
gaia_data.inp Symbolic_gaia_data
result.dat Symbolic_result Finally, our code with symbolic names looks like:
#PLEIADES - BOINC version
#Searching the Pleiades and computing the mean value and standard deviation of proper motion
import numpy as np
#loading gaia results (using numpy to read whole table)
#use defined symbolic name as file names when opening the files
r=np.loadtxt(fname="Symbolic_gaia_data", skiprows=1)
#constants are defined in an input config file
with open("Symbolic_config") as f:
line = f.readlines()[0].split(' ')
pmra0=float(line[0])
pmdec0=float(line[1])
#filtering the output from Gaia archive for objects with (pmra-pmra0)**2+(pmdec-pmdec0**2) < 5**2
#and saving results to numpy arrays
pm_ra=np.array([])
pm_dec=np.array([])
for row in r:
if (row[0]-pmra0)**2+(row[1]-pmdec0)**2 < 25.0:
pm_ra=np.append(pm_ra,row[0])
pm_dec=np.append(pm_dec,row[1])
#computing mean values and standard deviation using numpy
mean_pmra=np.mean(pm_ra)
mean_pmdec=np.mean(pm_dec)
stdev_pmra=np.std(pm_ra)
stdev_pmdec=np.std(pm_dec)
#saving results to output file
with open("Symbolic_result",'a') as f:
f.write(f'{mean_pmra},{mean_pmdec}\n{stdev_pmra},{stdev_pmdec}\n')IV. Compilation
Compilations should be performed on the target system on which the program is to run and run the appropriate compilers:
- Windows py2win
- MacOS py2app
- Unix pyinstaller
We will compile the code under Unix:
pyinstaller --onefile example.py -n pleiades_unixThe PyInstaller will create an executable file in ./dist catalogue. This file can be used in Gaia@Home.
V. Give name to the task
Click on "New task" on the web.gaiaathome.eu :
![]()
then enter project name:
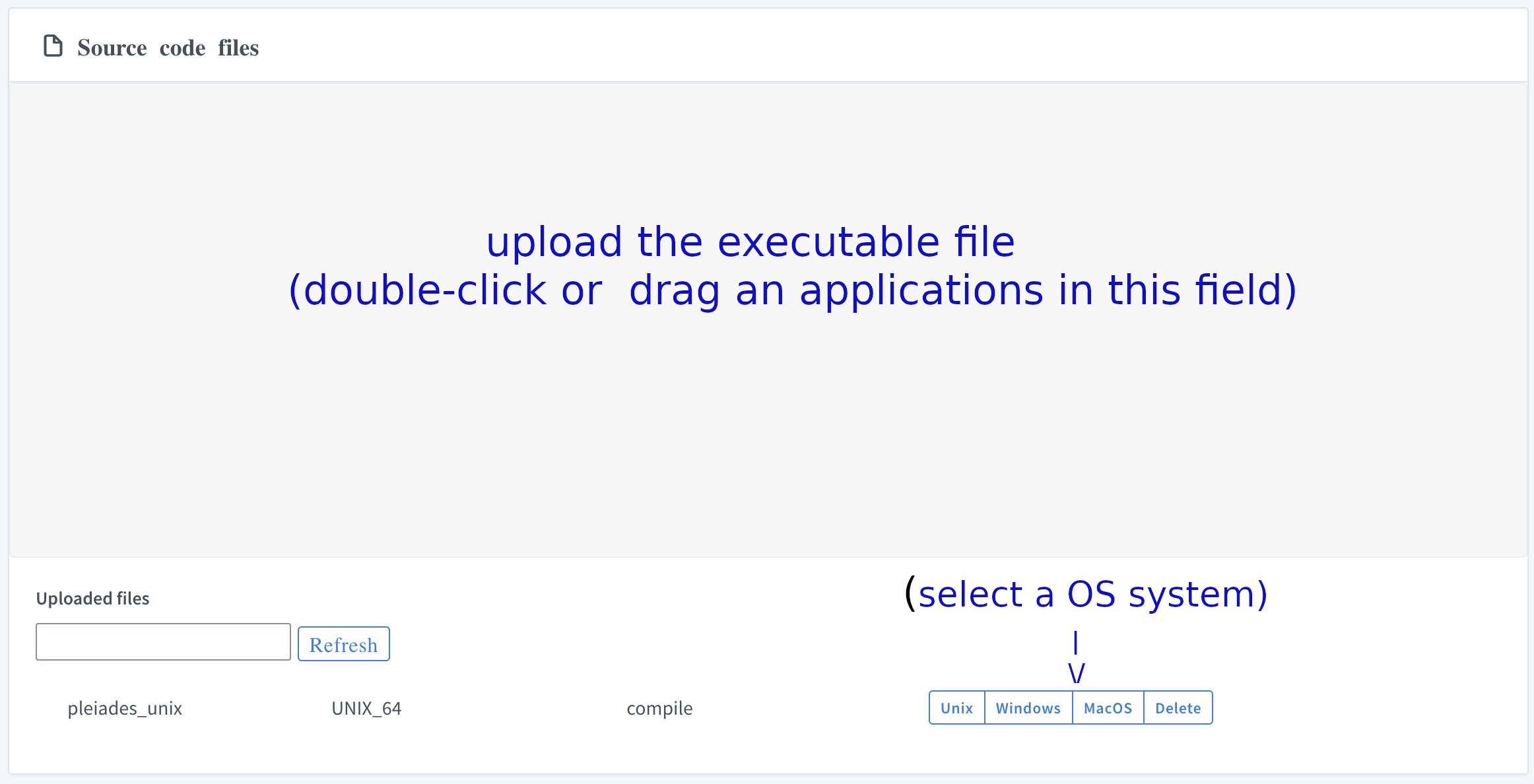
VI. Submit application
Upload executable application (pleiades_unix) to the system:
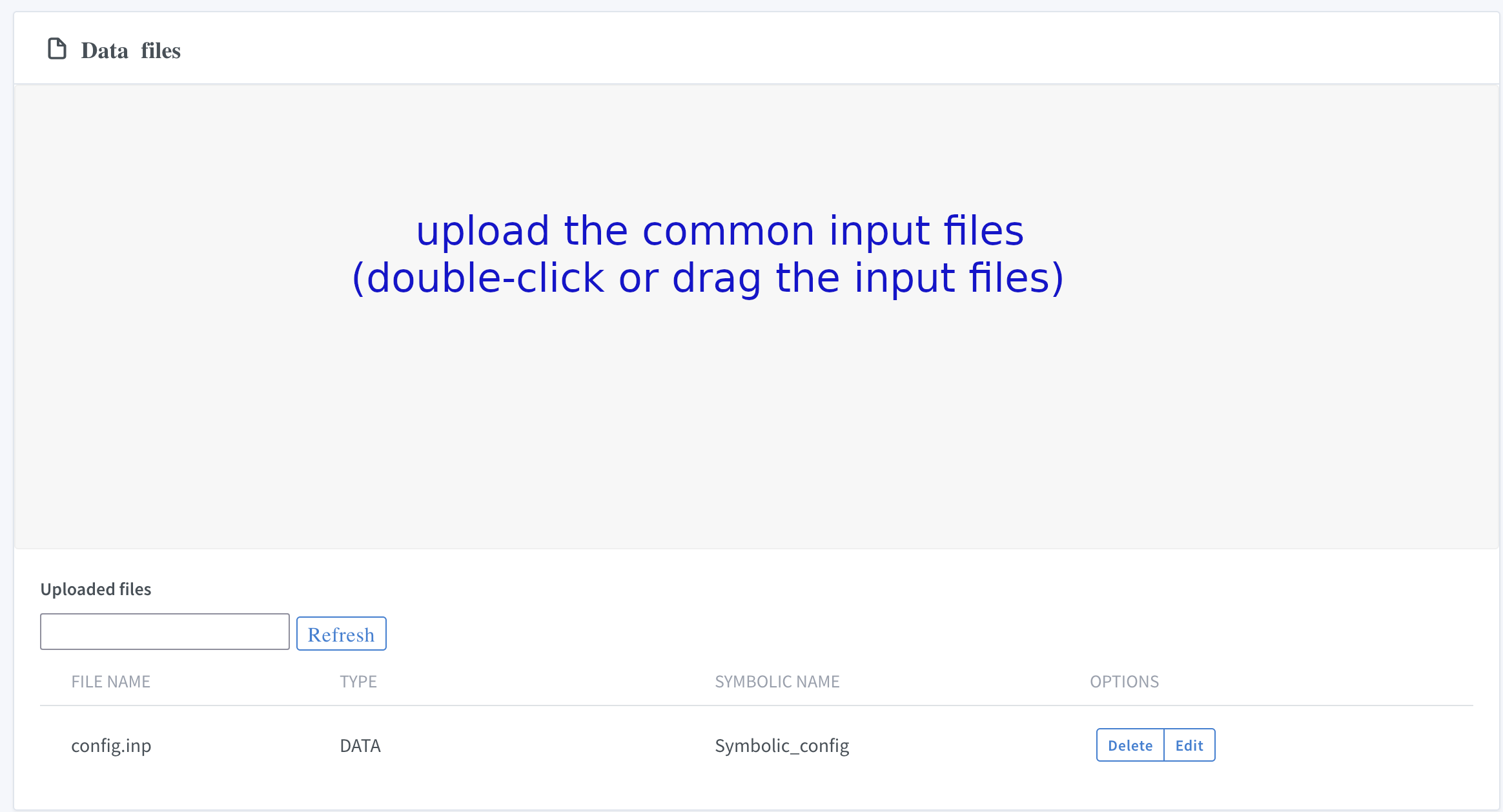
VII. Submit common input files
Upload common input files to the system and edit the symbolic name (optional):
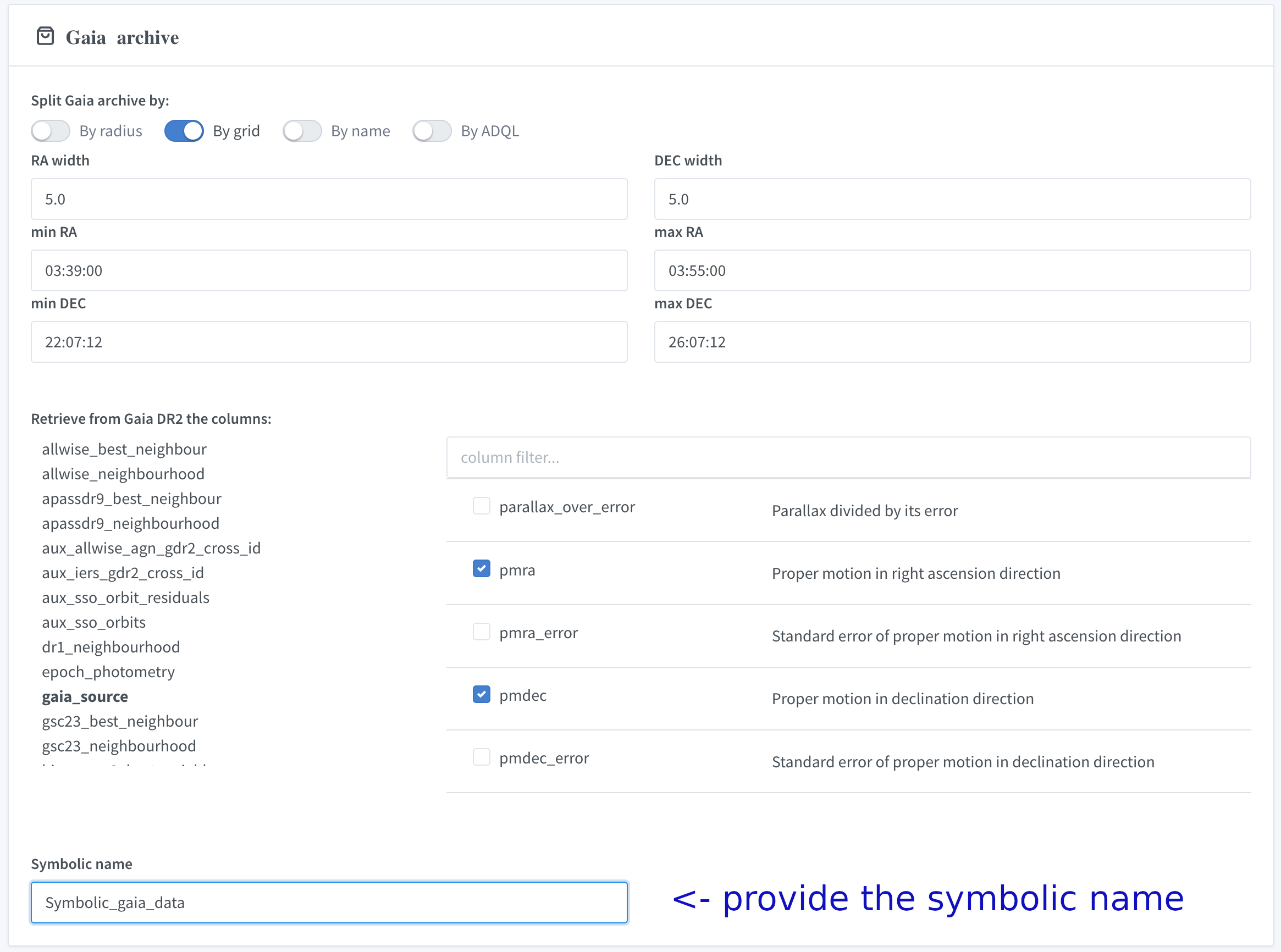
VIII. Prepare a query for the Gaia archive
Set the type and parameters of the Gaia archive query, specify the returned data fields and enter the symbolic name :
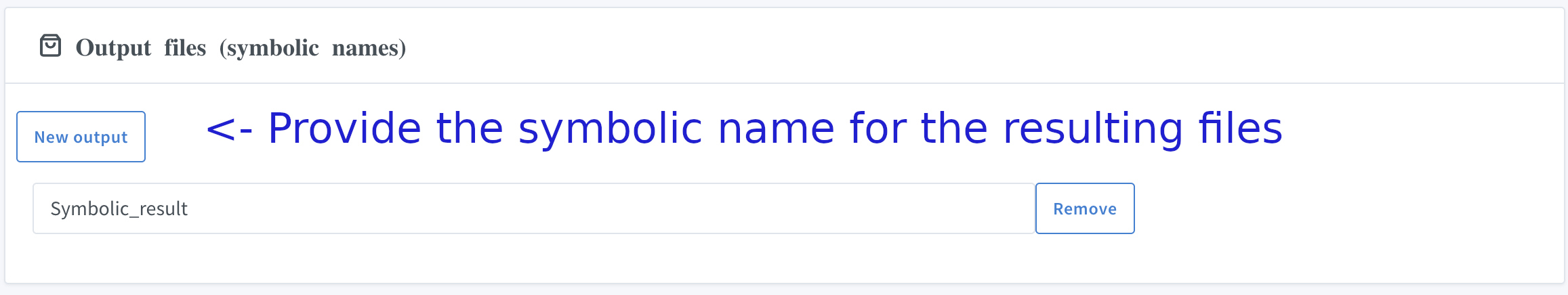
IX. Provide the names of the output files
Provide the names of the output files:
X. Start the calculations
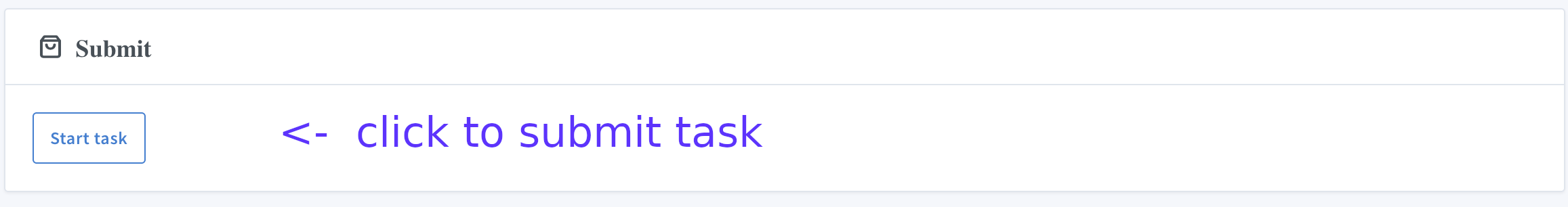
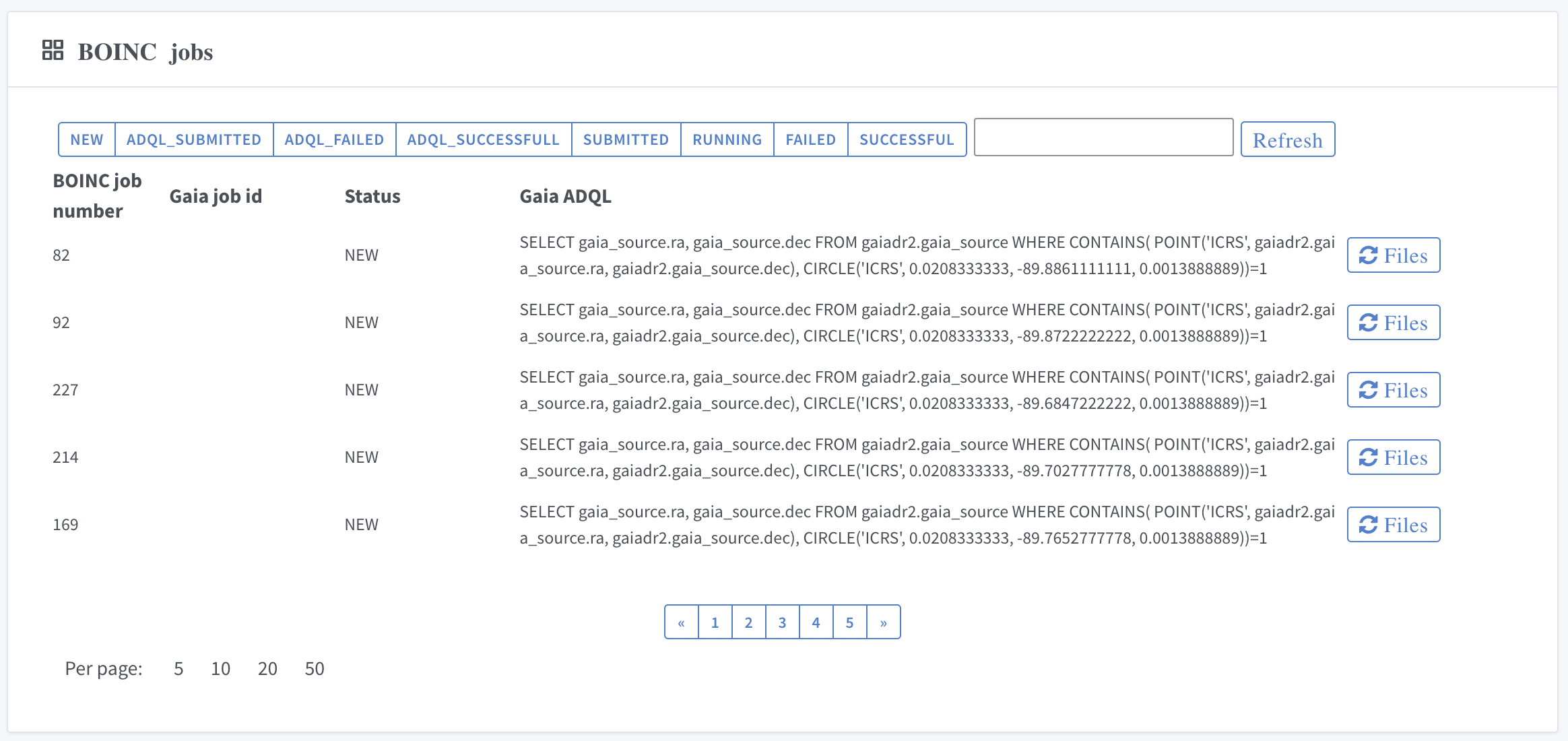
Press "Tasks" :

to see the list of tasks:
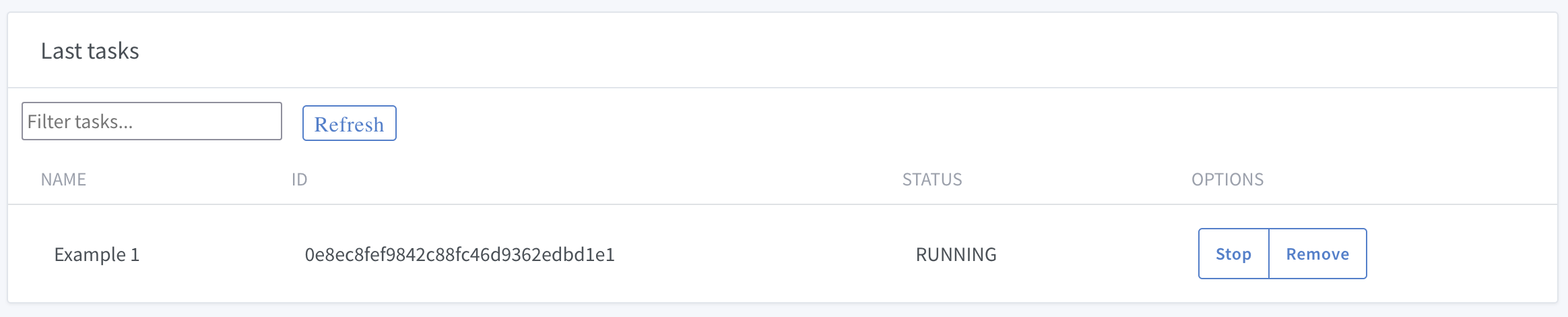
If you want to interrupt your calculations, you can stop or remove the tasks.
XII. Download results from completed jobs
By clicking on the task name you will get the details of the task:
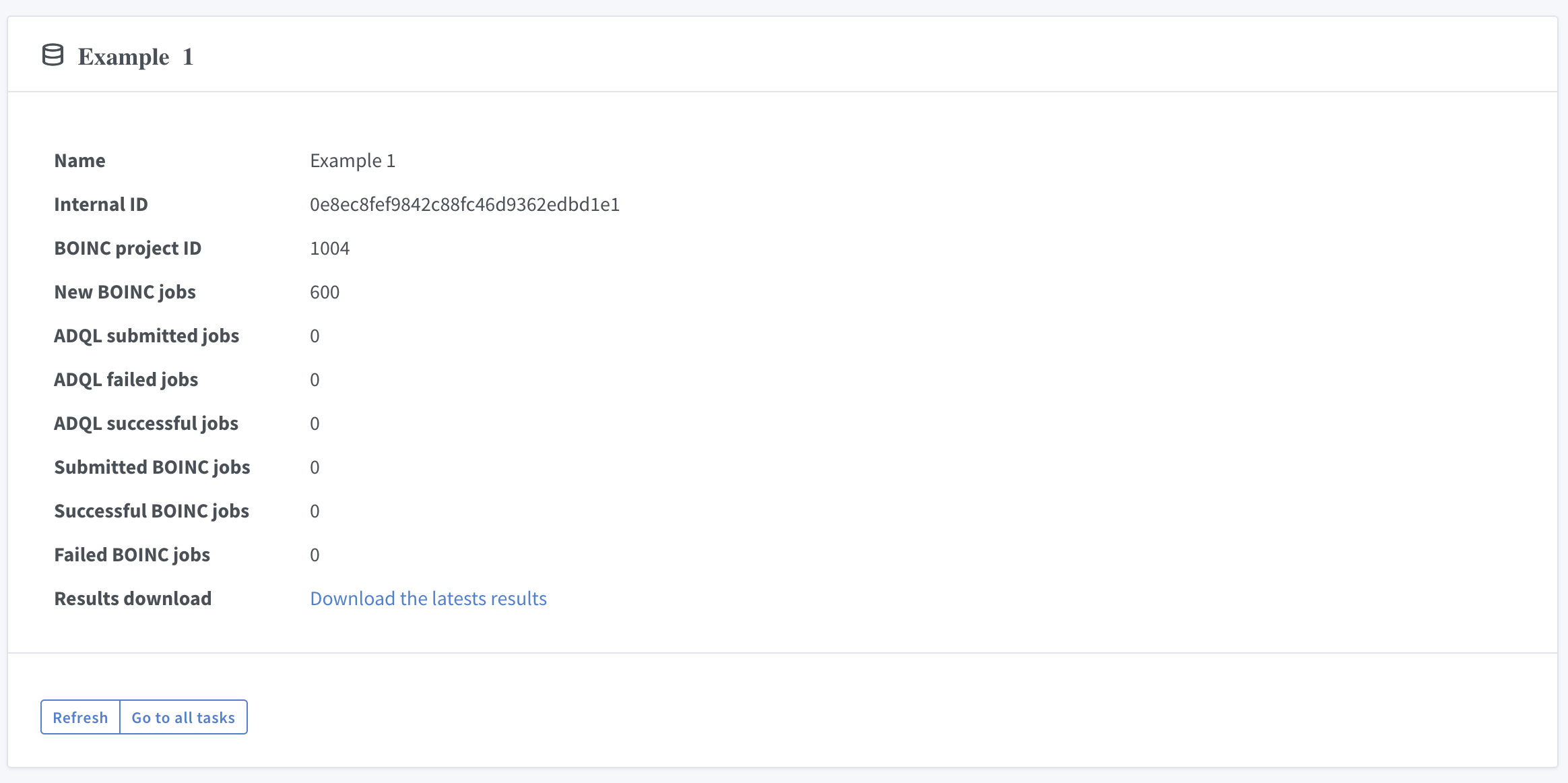
Link "Download the latests results" triggers download of received calculation results.
The system also provides detailed information on individual jobs: